Introduction: CD79B is a target of polatuzumab vedotin, an antibody-drug conjugate, which significantly improved the prognosis of both previously untreated and relapsed/refractory patients with diffuse large B-cell lymphoma (DLBCL). However, the biological and clinical significance of CD79B protein and gene expression have not been fully explored in DLBCL, thus we aimed at examining these relationships.
Methods: We retrospectively analyzed de novo DLBCL patients, who were diagnosed and received rituximab-based immunochemotherapy from 2008 through 2018 in the Okayama Hematology Study Group from Japan. Immunohistochemistry (IHC) staining was performed using a CD79B antibody (AT107-2), and protein expression was assessed based on H-score as described in a previous study (Sehn LH et al. JCO 2020), integrating with publicly available representative bulk RNA sequencing DLBCL datasets (BCC cohort from Ennishi D et al. JCO 2019 and NCI cohort from Schmitz R et al. NEJM 2018). We also performed CD8 and MHC class-I IHC to evaluate the tumor microenvironment. Gene expression profile-based cell-of-origin (COO) classification was performed including double-hit signature (DHITsig), recently renamed the dark zone signature (DZsig), using the NanoString DLBCL90 assay. In addition, simultaneous epitope and transcriptome measurement in single cells from lymphoid tissues was conducted.
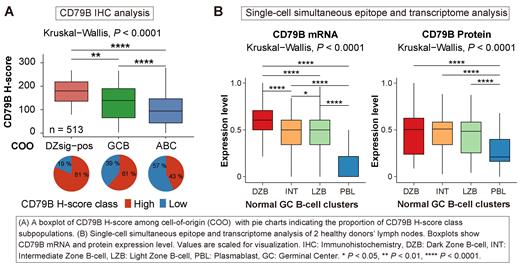
Results: CD79B IHC was evaluable in 576 cases. DLBCL90 assay classified the entire cohort into 293 ABC (50.9 %), 189 GCB (32.8 %), 31 DZsig-positive (5.4 %) and 63 unclassified (10.9 %). Furthermore, we dichotomized the cohort into 288 CD79B high cases and 288 CD79B low cases according to the median CD79B H-score. A dynamic range of CD79B protein expression was observed across COO, where ABC-DLBCL showed the lowest values followed by GCB-DLBCL and DZsig-positive-DLBCL, in ascending order (Kruskal-Wallis test, P < .00001; Figure A). Indeed, CD79B low cases were significantly enriched in ABC-DLBCL (58 %) compared to GCB-DLBCL (26 %) and DZsig-positive-DLBCL (2 %), respectively (Chi-squared test, P < .001). Consistently, we revealed that CD79B expression was the lowest in ABC-DLBCL compared to GCB-DLBCL and DZsig-positive-DLBCL at the transcriptomic level (Kruskal-Wallis test, P = .011 for BCC cohort and P = .022 for NCI cohort). In addition, we identified different CD79B staining patterns, composed of 433 with cytoplasmic pattern (75 %), 86 with membranous pattern (14.9 %), and 52 cases being IHC negative. These patterns significantly varied across COO (Fisher's exact test, P = .015) and CD79B H-score was the highest in the membranous pattern followed by the cytoplasmic pattern (Kruskal-Wallis test, P < .0001). Of note, the composition of CD8 positive T-cells in CD79B low tumors was significantly higher than that of CD79B high tumors (Wilcoxon rank sum test, P < .0001). However, MHC class-I expression was decreased in CD79B low cases compared to CD79B high (Wilcoxon rank sum test, P = .0002), suggesting an immune escape mechanism with downregulation of MHC class-I in the presence of cytotoxic T-cells, which is often seen in solid cancers. Significant association of CD79B expression with COO further prompted us to evaluate CD79B expression in normal germinal center B cells. Notably, the single-cell simultaneous epitope and transcriptome analysis (CITEseq, n = 2) and single-cell RNAseq analysis (n = 6) of reactive lymph nodes revealed that both CD79B gene and protein expression were the lowest in cells exhibiting plasmablastic signatures, followed by light zone and dark zone B cells (Kruskal-Wallis test, P < .0001; Figure B), supporting the relation of CD79B expression to COO subtype in DLBCL. Regarding prognostic impact, CD79B low group had significantly poorer prognosis in the entire DLBCL cohort (Log-rank test, P = .0005 for overall survival (OS) and P = .008 for progression-free survival (PFS)) and in ABC-DLBCL (Log-rank test, P = .003 for OS and P = .031 for PFS). Moreover, CD79B protein expression was significantly associated with OS after adjusting for International Prognostic Index in the entire DLBCL cohort (Cox regression model; P = .035).
Conclusion: Our study identifies distinct CD79B expression patterns across COO subtypes, with CD79B low cases enriched in ABC-DLBCL and demonstrating poorer prognosis, suggesting its potential as a prognostic marker and for targeted therapies.
Disclosures
Sunami:Chugai Pharma: Research Funding; Abbvie, Incyte, GlaxoSmithKline, Novartis, Pfizer, BeiGene, Kyowa Kirin, Ono, Otsuka and Chugai: Research Funding; Sanofi, BMS and Janssen: Honoraria, Research Funding. Hiramatsu:AbbVie, Novartis, Kyowa Kirin, Otsuka and Chugai: Honoraria; Sanofi, BMS and Janssen: Honoraria, Research Funding. Yoshida:Janssen, Eisai, Dainipponsumitomo, BMS, MeijiSeika pharma, NipponKayaku and Astrazeneca: Honoraria; Kyowa Kirin, Chugai: Honoraria, Research Funding. Kobayashi:Chugai, Sanofi, Asahi Kasei Pharma, Janssen and Meiji Seika Pharma: Honoraria. Sato:Chugai, Kyowa Kirin and Takeda,: Honoraria; Daiichi Sankyo: Current holder of stock options in a privately-held company, Honoraria. Maeda:Eisai Pharmaceutical Co., Ltd.: Honoraria; Takeda Pharmaceutical Co., Ltd: Honoraria; Kyowa Kirin: Honoraria; AstraZeneca: Research Funding; Chugai Pharmaceutical Co., Ltd.: Honoraria, Research Funding; Astellas: Honoraria, Research Funding; Nippon Shinyaku Pharmaceutical Co., Ltd.: Honoraria, Research Funding; Otsuka Pharmaceutical: Honoraria; Novartis Pharma: Research Funding, Speakers Bureau. Scott:Abbvie, AstraZeneca, Incyte: Consultancy; Janssen and Roche: Research Funding. Ennishi:BMS: Honoraria; Kyowa Hakko Kirin Co., Ltd.: Honoraria; Ono Pharmaceutical: Honoraria; Janssen: Honoraria; Incyte: Honoraria; Eisai: Honoraria, Research Funding; Chugai: Honoraria, Research Funding; Nippon Shinyaku Co.: Honoraria, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal